For nearly 30 years, my principal interest has been the role of rearrangements of large segments of the genome, i.e. chromosome translocations, duplications, deletions, and inversions, in common cancers such as breast cancer (reviewed in Edwards, J Pathol 2010;220:244-54).
These have proved particularly difficult to study in common cancers, but they are abundant and have powerful effects, particularly where they create fusion genes—genes formed by the fusion of two genes, like the BCR-ABL fusion of chronic myeloid leukaemia. We estimated an average breast cancer may express around 10 fusion genes (Edwards & Howarth, Breast Cancer Research 2012;14, 303).
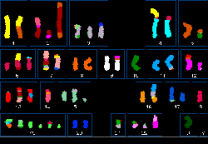
A good starting point to understand genome rearrangements at the largest scale is the multicolour fluorescent ‘SKY’ images like the one shown here, which outline the major chromosome translocations in a tumour. Our catalogue of cell line karyotypes is now at https://web.expasy.org/cellosaurus/pawefish. Following this, we used arrays of DNA sequences to map the boundaries of the chromosome segments in these images. We found that reciprocal translocations often have tens of kilobases of common sequence duplicated onto both product chromosomes, leading us to propose that chromosome translocations are formed at replication bubbles (Howarth et al., 2011).
We moved on to massively parallel DNA and RNA sequencing, both in breast cancer and oesophageal adenocarcinomas, in collaboration with Professor Carlos Caldas and Rebecca Fitzgerald in the Department of Oncology, respectively. Among the highlights have been using knowledge of genome evolution in breast cancers to distinguish early and late mutations (Newman et al., 2013); and finding that the oesophageal adenocarcinomas have an exceptionally high incidence of novel LINE-1 mobile element insertions, which contribute to genetic change but remain largely uncharacterized (Paterson et al BMC Genomics 2015;16:473). The current focus is on detecting fusion genes that may be drug targets, such as NRG1 fusions (Howarth et al., 2021).
We have also worked on methods for mapping genomes that rely on sampling: parts of the genome that are close together always turn up in the same samples. We showed that the linear form of this approach, HAPPY mapping, could be used to map cancer rearrangements (Pole et al, Nucleic Acids Res 2011; 39, e85). Generalizing the approach to 3D led to the development of the GAM method for mapping the arrangement of DNA in the nucleus (Beagrie et al., 2017).
Key Publications
- Edwards PAW. Re-interpreting tumour behaviour and the tumour microenvironment as normal responses to tissue disorganisation. J. Pathol. 2023; 260, 1–4
- Howarth KD, Mirza T, Cooke SL, Chin S-F, Pole JC, Turro E, Eldridge MD, Garcia RM, Rueda OM, Boursnell C, Abraham JE, Caldas C, Edwards PAW. NRG1 fusions in breast cancer. Breast Cancer Res. 2021;23:3.
- RA Beagrie, A Scialdone, M Schueler, DCA Kraemer, M Chotalia, SQ Xie, M Barbieri, I de Santiago, L-M Lavitas, MR Branco, JF, JDostie, L Game, N Dillon, PAW Edwards, M Nicodemi, A Pombo Complex multi-enhancer contacts captured by Genome Architecture Mapping (GAM) Nature 2017;543 (7646),519-524.
- Newman S, Howarth KD, Greenman CD, Bignell GR, Tavaré S, Edwards PAW. (2013) The relative timing of mutations in a breast cancer genome. PLoS ONE. 2013;8:e64991.
- Howarth KD, Pole JCM, Beavis JC, Batty EM, Newman S, Bignell GR, Edwards PAW. Large duplications at reciprocal translocation breakpoints might be the counterpart of large deletions and could arise from stalled replication bubbles. Genome Research 2011; 21, 525-534.