Our Research
Bacterial toxin export and multidrug efflux
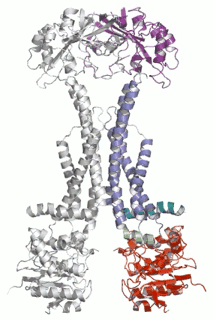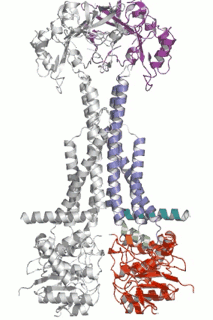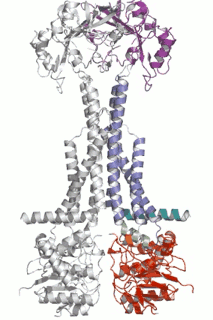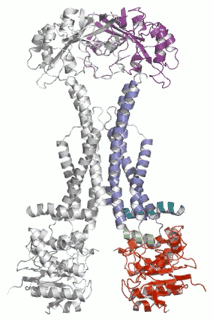
Diverse molecules, from small antibacterial drugs to large protein toxins, are exported directly across the membranes of Gram-negative bacteria. This is achieved by the reversible interaction of a two component inner membrane (IM) translocase, which provides substrate specificity and energy, with a protein of the TolC 'channel-tunnel' family anchored in the outer membrane (OM) that spans the periplasmic space. Having determined the high-resolution structure of TolC, we are using biochemical and biophysical methods in combination with structural biology to probe channel-tunnel dynamics and to identify potential inhibitory agents. In parallel, we are investigating the structure and function of inner membrane translocase components and their interactions with TolC.
Action of Salmonella effector proteins during mammalian cell entry
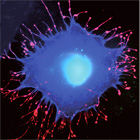
Salmonella species continue to cause intestinal disease. Critical to the onset of infection is the ability of the bacteria to enter cells of their mammalian host, from where they replicate and spread. To achieve this, the bacteria deliver a cocktail of effector proteins into the host cell, which subvert signal transduction pathways and stimulate cytoskeletal rearrangements, and trick the normally dormant target cells into engulfing the pathogen. Our goal is to decipher this multifactorial cross-talk between Salmonella effectors and the eukaryotic cellular machinery using a combination of molecular and cellular biology.
Exploiting pathogenic E.coli to model transmembrane receptor signalling
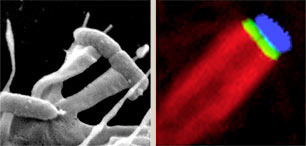
Enteropathogenic and enterohaemorrhagic E.coli, which both cause severe diarrhoeal disease, can adhere to mammalian intestinal cells and induce reorganization of the actin cytoskeleton into 'pedestal-like' pseudopods beneath the extracellular bacteria. As pedestal assembly is triggered by E.coli virulence factors that mimic several host cell-signalling components, such as transmembrane receptors, their cognate ligands and cytoplasmic adaptor proteins, it can serve as a powerful model system to study eukaryotic transmembrane signalling. We are exploiting these bacterial factors as versatile tools to probe central cellular signalling pathways.
Deciphering cellular signalling to the cytoskeleton
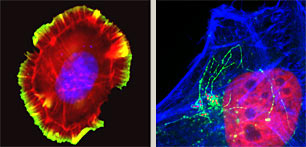
Distinct from ongoing experiments analyzing pathogenesis, we are exploiting effector activities to establish diverse new projects that address complex outstanding questions in cell biology. One such intriguing problem is understanding how cells move. Cells navigate by assembling projections to extend their plasma membrane forward while co-ordinately retracting analogous structures at the opposing edge. We are establishing new techniques to analyze the underlying protein complexes and lipid-mediated signalling using in vitro reconstitution and biomimetic approaches.
Professor Vassilis Koronakis
Principal Investigator
Group Members