Our Research
Bacterial motility & flagella biogenesis Pathogenic bacteria are often motile and their ability to swim through liquids and swarm over surfaces facilitates pathogen-host interactions and colonization of nutrient–rich environments. Many bacteria build complex rotary nanomotors, called flagella, which drive swimming and swarming motility. A remarkable feature of flagella is that they are essentially self-assembling. Thousands of flagellar structural subunits are exported across the cell membrane by dedicated type III export machinery located at the base of each flagellum. Subunits then transit through a narrow channel at the core of the nascent flagellum to reach the assembly site at the tip of the structure, up to 20 μm away from the cell surface. Our work aims to uncover the molecular mechanisms underpinning flagellum assembly, focusing on the interactions between flagellar structural proteins and components of the membrane export machinery. Funded by a BBSRC project grant.
A chain mechanism for bacterial flagellum growth (see Nature 504:287-290). Flagella on the surface of the bacterial cell (inset top right, Salmonella cell membrane stained green and flagella stained red) comprise three contiguous substructures - the basal body, hook and filament - that are assembled sequentially. A central channel runs through these substructures to the flagellum tip.The basal body houses the flagellar export machinery (A) and the rod, which extends from the inner membrane (IM) and crosses the periplasm, peptidoglycan (PG) cell wall and outer membrane (OM). The cell surface hook is a flexible universal joint that connects the external flagellum filament to the basal body.
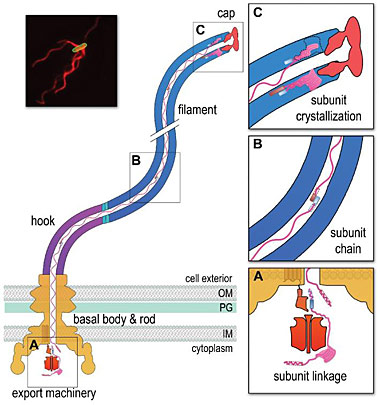
(A) Subunits of the rod, hook and filament link head-to-tail at the cytoplasmic membrane export machinery. Subunits are first unfolded by the export ATPase complex (red). Rod and hook subunits then dock at the FlhB export gate (orange). The N-terminal helix of the docked subunit is then captured by the free C-terminal helix of an exiting subunit in the flagellum channel. (B) Sequential subunits are linked head-to-tail in a chain by juxtaposed terminal helices forming parallel coiled-coils. The resulting chain of unfolded subunits is connected through the flagellum central channel to the distal tip of the growing structure. (C) The subunits in the chain transit from the gate to the tip. Subunits fold and incorporate into the flagellum tip beneath cap foldases. Subunit folding and/or crystallization not only provides a strong anchor at the flagellum tip, it also shortens the subunit chain in the channel thus exerting a pulling force on the next subunit at the export gate, pulling it from the gate. The pulling force then drops rapidly as the new unfolded subunit enters the channel. This process repeats for each subunit captured into the chain.
Bacterial nucleoid structure & gene expression The bacterial nucleoid is a large dynamic structure containing DNA, RNA and proteins. To fit into the cell the nucleoid must be compacted extensively, and must fold in ways that allow genetic information to be replicated and transcribed. This is especially important in bacteria as their survival depends on their ability to adapt rapidly to the external environment by modulating gene expression and growth rate. We aim to understand how the nucleoid folds and how this folding influences transcription. Nucleoid folding is orchestrated, in part, by specialized DNA-binding proteins called nucleoid associated proteins (NAPs). We have mapped the genomic binding sites of several NAPs - including Fis, HNS, IHF and HU - in the model bacterium E. coli K12. We have also determined the effects of NAP binding on both gene expression and genome-wide binding of RNA polymerase. We are now building on this work to study how NAPs influence long-range DNA interactions and the 3D structure of the nucleoid. In collaboration with Pietro Cicuta, Department of Physics.
Dr Gillian Fraser
Principal Investigator
|
|


