Biography
I received my BSc cum laude and with General Honours from the University of Miami, majoring in Microbiology & Immunology with a double minor in Chemistry and Biology. I earned my PhD and dual Masters degrees from Columbia University's Department of Microbiology & Immunology in New York City, where I worked with Professor David Shore on transcriptional silencing and heterochromatin assembly in the budding yeast, Saccharomyces cerevisiae.
After being awarded a Fellowship from the Imperial Cancer Research Fund (now the Francis Crick Institute), I moved to London for post-doctoral research. I studied KSHV-encoded cyclins with Dr Nic Jones and cell cycle regulation with Dr Gordon Peters. My discovery of the critical role ubiquitin ligases play in regulating G1 phase kinases in KSHV-driven malignancies led to funding from the Association for International Cancer Research to continue this work with Professor Chris Boshoff at the Wolfson Institute for Biomedical Research at University College London.
In 2005, I was awarded a Research Fellowship to establish my independent research group in the Department of Pathology at the University of Cambridge. I am now Professor in Cellular and Molecular Biology and serve as Head of the Department. As a Fellow of Clare College, Cambridge, I am Director of Studies for Pathology and Genetics and the Postgraduate Admissions Tutor.
Research
Research overview
How organisms achieve their intriguing biological complexity with an amazing ability to respond to environmental stress and infections is a fascinating and perplexing question. One strategy is through the action of enzymes that modify proteins to rapidly change protein behaviour and function. Ubiquitin ligases are a family of enzymes my lab investigates which are integral to this. They tag their substrates with a 76 amino acid tag, called ubiquitin. This modification can change a protein’s stability, localisation and trafficking within the cell. Ubiquitin signalling has different consequences in different cell types and under different conditions. My lab studies this signalling in normal and pathological conditions to understand the mechanisms of a wide range of diseases, like Parkinson’s disease, cancer, male sterility and anaemia. We aim to exploit the exquisite specificity of ubiquitin ligase enzymology to invent novel therapeutics.
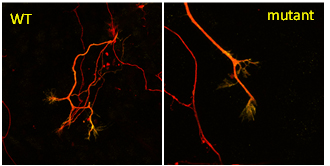
Neuroprotective roles of Fbxo7/PARK15
Parkinson’s disease is the second most common neurodegenerative disease, affecting more than 6 million people worldwide, where the majority of cases are sporadic and approximately 10% of cases are inherited. In 2008, the first case of Parkinsonian pyramidal syndrome was linked to a mutation in FBXO7, and since then many other pathological recessive mutations have been identified. We are using cell lines, 2D and 3D dopaminergic neurons, and mouse models to identify which cellular pathways will rescue the ensuing neuronal cell death caused by the loss of Fbxo7.
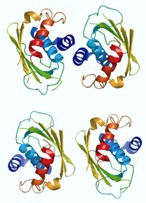
Fbxo7 and PI31 as proteasome regulators
In studying the structure and functional domains of Fbxo7, we discovered it also contains an FP (Fbxo7-PI31) dimerisation domain that enables its homo-dimerization and hetero-dimerisation with the proteasome regulator, PI31/PSMF1. These proteins share a similar organisational structure, both contain an FP domain and a PRR domain, and reduced Fbxo7 levels often correlates with lower PI31 levels. Multiple global proteomic profiling and interactome studies have demonstrated the association of Fbxo7 and PI31 with each other and the proteasome. Our goal is to understand how Fbxo7 and PI31 coordinate their mutual regulation of the proteasome in the multiple different cell types where Fbxo7 deficiency shows a pathological phenotype.
F-box proteins in cancer
F-box proteins, including Skp2 and Fbxw7, control the ubiquitin-mediated degradation of key cell cycle regulators, like p27, cyclin E and Myc. We aim to assess whether when tumours present with mutations in F-box protein genes, whether these can be used as biomarkers for deciding on chemotherapeutic treatments for patients. We collaborate with AstraZeneca in understanding the tumour biology of subsets of breast cancers with regard to their ubiquitin signalling networks.
Targeting ubiquitin ligases and ubiquitinated proteins with novel biotherapeutic approaches
Recent advances in converting small molecule ligands into protein-targeting chimeric (PROTACs) molecules demonstrate the power of the ubiquitin-proteasome system to selectively degrade proteins within the cell. Ubiquitin ligases are remarkably specific enzymes that target their substrates via their F-box domain containing subunits. We want to exploit the ability of F-box proteins to bind their substrate to identify protein-protein interacting sites to convert into biological Protacs. We use camelid-derived antibodies, known as nanobodies, to test and disrupt the interactions between ubiquitin ligases and their substrates. Our goal is to define the protein-docking sites for ligands in difficult-to-target oncogenes.
Group Members:
Gudrun Agnarsdottir, Sara Al Rawi, Hanna Bjone, Ting Deng, Finn Snow, Daniel Tikhorimov, Sophie Willis, Libby Wiseman, Min Zhang
Publications
-
Al Rawi S, Tyers P, Barker RA, Laman H. Restoration of Fbxo7 expression in dopaminergic neurons restores tyrosine hydroxylase in a mouse model of Parkinson’s disease. biorxiv 2024 Oct. Link.
-
Al Rawi S*, Simpson L*, Agnarsdóttir G, McDonald NQ, Chernuha V, Elpeleg O, Zeviani M, Barker RA, Spiegel R^, Laman H.^ Study of an FBXO7 patient mutation reveals Fbxo7 and PI31 co-regulate proteasomes and mitochondria. FEBS J 2024 Feb. Link.
-
Harris R, Yang M, Schmidt C, Royet C, Singh S, Natarajan A, Morris M, Frezza C, Laman H.Fbxo7 promotes Cdk6 activity to inhibit PFKP and glycolysis in T cells. J Cell Biol. 2022 Jun 7. doi: 10.1083/jcb.202203095. Link.
-
R. Harris, S.J. Randle & H Laman. Analysis of the FBXO7 promoter reveals overlapping Pax5 and c-Myb binding sites functioning in B cells. 2021. Biochem Biophys Res Comm. Link
-
Licchesi JDF, Laman H, Ikeda F, Bolanos-Garcia VM. Front Physiol. 2020 Nov 26;11:621053. Link.
-
Mason BJ & Laman H. The FBXL family of F-box proteins: variations on a theme. Open Biol. 2020 Nov;10(11):200319. Link
-
Licchesi JDF, Laman H, Ikeda F, Bolanos-Garcia VM. E3 Ubiquitin Ligases: From Structure to Physiology. Front Physiol. 2020. 26 November 2020 Link
-
Spagnol V, Oliveira CAB, Randle SJ, Passos PMS, Correia CRSTB, Simaroli NB, Oliveira JS, Mevissen TET, Medeiros AC, Gomes MD, Komander D, Laman H, Teixeira FR.The E3 ubiquitin ligase SCF(Fbxo7) mediates proteasomal degradation of UXT isoform 2 (UXT-V2) to inhibit the NF-kappa B signaling pathway. Biochimica et Biophysica Acta. 2020 Sep 30;1865(1):129754. doi: 10.1016/j.bbagen.2020.129754. Link
-
Rathje CC, Randle SJ, Al Rawi S, Skinner BM, Rajamundar A, Nelson DE, Johnson EEP, Bacon J, Vlazak M, Affara NA, Ellis PJ, Laman H.A conserved requirement for Fbxo7 during male germ cell cytoplasmic remodelling. Front Physiol. 2019. Oct 10;10:1278. doi: 10.3389/fphys.2019.01278. Link.
-
Mason B, Flach S, Teixeira, FR, Manzano Garcia R, Rueda OM, Abraham JE, Caldas C, Edwards PAW. Laman H. Fbxl17 is rearranged in breast cancer and loss of its activity leads to increased global O-GlcNAcylation. Cell Mol Life Sci. 2019 Sept 27. Link.
- SRW Stott†, SJ Randle†, S al Rawi, PA Rowicka, R Harris2, B Mason, J Xia, JW Dalley, RA Barker, H Laman. Loss of FBXO7 results in a Parkinson’s-like dopaminergic degeneration via an RPL23-MDM2-TP53 pathway. J Pathol. 2019 Oct;249(2):241-254. Link.