Submitted by Livia Harriman on Wed, 13/12/2023 - 12:16
Our recent study delving into bacterial pathogens, particularly non-typhoidal Salmonella and Listeria monocytogenes, has uncovered striking differences in their translational regulation mechanisms.
These pathogens, responsible for millions of infections and over 230,000 annual fatalities, deploy common strategies to adhere to, invade, and establish infections within host cells.
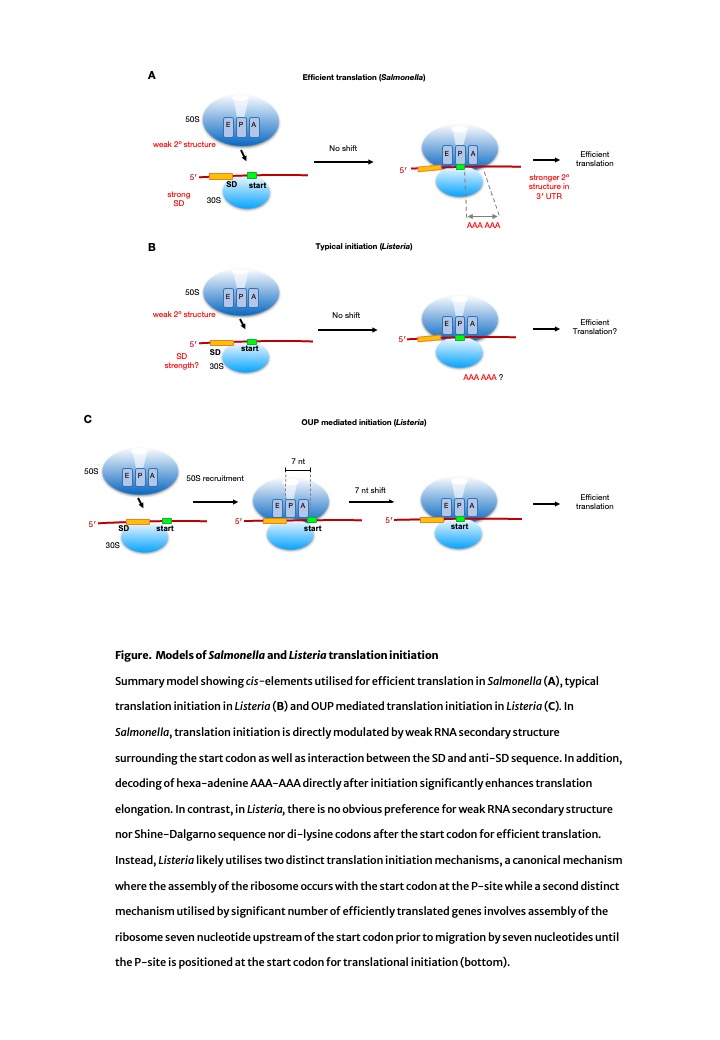
Utilizing cutting-edge high-definition translatomic and transcriptomic technologies, our study has unveiled distinct translational control mechanisms between these two divergent bacteria. In Salmonella, efficient protein synthesis depends on strong Shine-Dalgarno sequences and/or weak secondary structures, aligning with highly established modes observed in Gram-negative bacteria like E. coli. In contrast, Listeria, a highly divergent Gram-positive bacterium, defies these norms and reveals a novel 'Out-of-frame Upstream Peak' (OUP)-mediated initiation of protein synthesis mechanism. Bacterial translation findings shed light on the intricate translational nuances governing these major pathogens. Our discovery of the OUP-mediated initiation in Listeria, responsible for 40% of its protein production, unveils a depth to bacterial translation initiation beyond prior expectations!
Read the paper: The distinct translational landscapes of gram-negative Salmonella and gram-positive Listeria by Owain J. Bryant#, Filip Lastovka#, Jessica Powell and Betty Y-W Chung 2023 Nature Communications https://www.nature.com/articles/s41467-023-43759-1