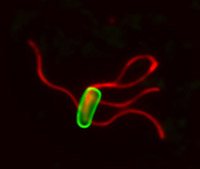
Bacteria are the smallest of living cells, and many like Salmonella and E. coli cause infection and disease. To respond to their environment and swim towards nutrients or away from danger, bacteria (green cell in photo) build helical propellers called flagella (red) on their surface.
Both biologists and physicists have long found flagella fascinating as they illustrate beautifully how complex biological structures are constructed to operate as tiny ‘nanomachines’ on the cell surface.
 During assembly of each flagellum, thousands of subunit ‘building blocks’ destined for the growing structure are made inside the cell, then unfolded and exported across the cell membrane. Like other biological functions, unfolding the subunits consumes energy generated by machines in the living cell. But then the subunits pass into a channel at the centre of the growing flagellum on the outside of the cell, and must transit a substantial distance to the flagellum tip where they assemble one after another into the structure. In this way the flagellum grows at a constant rate to several times the length of the cell.
During assembly of each flagellum, thousands of subunit ‘building blocks’ destined for the growing structure are made inside the cell, then unfolded and exported across the cell membrane. Like other biological functions, unfolding the subunits consumes energy generated by machines in the living cell. But then the subunits pass into a channel at the centre of the growing flagellum on the outside of the cell, and must transit a substantial distance to the flagellum tip where they assemble one after another into the structure. In this way the flagellum grows at a constant rate to several times the length of the cell.
The mystery has been how are the flagellar subunits passed down the long channel far outside the cell, where, there is no discernable energy source to propel them ?
This perplexing problem of flagellum growth has been unraveled by the findings of Dr Lewis Evans et al published in the journal Nature.
The Wellcome Trust-funded research, led by Dr Gillian Fraser and Professor Colin Hughes in the University of Cambridge, Department of Pathology, has uncovered a simple and elegant mechanism that allows constant growth of the flagellum outside the cell by harnessing the entropic force generated by the unfolded subunits themselves. The scientists showed that subunits bound at the base of the flagellum can be sequentially captured by head-to-tail linkage into a subunit chain that is pulled out through the entire length of the flagellum channel, powered by each subunit refolding and incorporating into the growing structure at the tip. Analysis performed by Professor Eugene Terentjev in the University’s Cavendish Laboratory confirmed that the pulling force generated by the chain of subunits adjusts automatically with each round of refolding and incorporation at the tip, so that continuous subunit transit is maintained at a constant rate. This mechanism describes how large proteinaceous structures can be assembled beyond the surface of living cells, and throws new light on how polymers move through channels, a classic problem in biology and physics.
This study is part of research carried out in the Tennis Court Road Microbiology Unit of Professors Hughes and Koronakis and Dr Fraser, supported for 28 years by the Wellcome Trust and UK Research Councils. Their work integrates molecular, cellular and structural biology approaches to understand the behaviour of Salmonella, E.coli and other pathogenic bacteria, especially the mechanistic basis of motility, virulence and multidrug resistance.

